 Another tool from the A* Vienna software team
Another tool from the A* Vienna software team
Pyckles is a super simple, light-weight interface to the Pickles (1998) catalogue of stellar spectra
pip install pyckles
Contents:
Warning
07.07.2020 Update to Brown et al. (2014) catalogue
Some referencing discrepancies were found in the Pyckles version of the Brown+ (2014) galaxy spectra catalogue. Pyckles will automatically re-download the catalogue if there is nothing in the astropy cache.
You can clear the astropy cache using
astropy.utils.data.clear_download_cache()
Warning
July 2022: The downloadable content server was retired and the data migrated to a new server.
Pyckles v0.2 and above have been redirected to a new server URL.
For older versions, please either upgrade to the latest version (pip install --upgrade pyckles), or follow these instructions to update the server URL in the config file.
Which spectra are available¶
Note
The package was originally intended only for the Pickles catalogue, but it now also has access to the Brown (2014) galaxy spectra catalogue
To list which catalogues are available, call get_catalog_list:
>>> import pyckles
>>> pyckles.get_catalog_list()
<Table length=2>
name type filename
str7 str7 str19
------- ------- -------------------
pickles stellar pickles98_full.fits
brown galaxy brown14_full.fits
To access any of the spectra in the library, we need to create a
SpectralLibrary object using the name of one of the available libraries:
>>> spec_lib = pyckles.SpectralLibrary("pickles")
To see a list of all the spectra in the catalogue, use the available_spectra
attribute:
>>> spec_lib.available_spectra
<Column name='name' dtype='str5' length=131>
A0I
A0III
A0IV
...
K2III
K3III
K4III
Accessing spectra¶
The spectra can be accessed both as items and as attributes of the
SpectralLibrary.
As an item:
>>> a0v = spec_lib["A0V"]
or as an attribute:
>>> a0v = spec_lib.A0V
Note
If there there is a space in the name, the attribute call will not
work - use the item call: spec_lib["my spec"]
By default Pyckles returns spectra as astropy.fits.BinTableHDU objects,
however depending on the use case, Pyckles can return spectra in any one of the
following formats. To change the way spectra are returned, we need to set the
meta parameter in the SpectralLibrary object:
>>> spec_lib.meta["return_style"] = "synphot" # default is "fits"
Acceptable settings are:
fits: returns spectra asfits.BinTableHDUobjects.wavelengthandfluxinformation is contained in the.dataattribute of theHDUsynphot: returns the spectra assynphot.SourceSpectrumobjectsquantity: returns the spectra as a tuple of twoastropy.Quantityarray objects in the format(wavelength, flux)array: returns the spectra as a tuple of twonumpy.ndarrayobjects in the format(wavelength, flux). The units here are those which were used in the FITS library (normallyangstromanderg s-1 cm-2 angstrom-1)
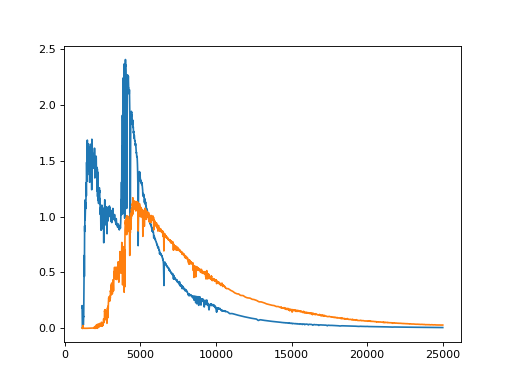
from matplotlib import pyplot as plt
import pyckles
spec_lib = pyckles.SpectralLibrary("pickles")
plt.plot(spec_lib.A0V.data["wavelength"], spec_lib.A0V.data["flux"])
plt.plot(spec_lib["G2V"].data["wavelength"], spec_lib["G2V"].data["flux"])
Adding libraries¶
If you have a spectral library that you would like added to pyckles, please
open an issue on github and we can talk about about adding in.
Library file formatting¶
Libraries are stored in a single FITS files with a contents table in the first extension, and the spectra in further BinTableHDU extensions
Extension |
HDU type |
Contents |
0 |
PrimaryHDU |
|
1 |
BinTableHDU |
Index table mapping spectra to extensions |
[1+n] |
BinTableHDU |
[n additional tables of spectra indexes] |
X |
BinTableHDU |
First of m spectra |
X+m |
BinTableHDU |
m-th spectra |
Index table extensions¶
The first extension should contain a table with (at a minimum) the following columns:
ext |
name |
X |
A0V |
X+1 |
A0III |
… |
… |
X+m |
M9V |
Additional spectra index tables can be included, but this means that all index
tables must be updated to keep the extension column pointing to the correct
spectra. For example in the pickles library, ext 1 indexes all the spectra
contained in the original Pickles (1998) library, however for ease of use, I
have added a further extension index in ext 2 which only includes the main
sequence stars. Additionally ext 3 contains a table with physical properties
of each of the spectral types, such as temperature and evolutionary stage. The
actual spectra are only contained onwards from ext 4.
Spectra extensions¶
The spectra are simple BinTableHDU objects containing 2 columns: wavelength
and flux.
wavelength |
flux |
… |
… |
If astropy is used to create these tables the headers will be
taken care of. If not, the following header information should be included:
TTYPE1 = 'wavelength'
TFORM1 = 'D '
TUNIT1 = 'Angstrom'
TTYPE2 = 'flux '
TFORM2 = 'D '
TUNIT2 = 'Angstrom-1 cm-1 erg s-1'
where the TUNITn keywords are in readable by astropy.units.
Contact¶
If you find any bugs with the code, or have suggestions for how the code can be improved, please open an issue on github: https://github.com/AstarVienna/Pyckles
If you are feeling adventurous, you’re more than welcome to play with the code and submit a pull request with any changes you deem necessary.
We’re more than happy for this to be incorporated into the astropy framework, if anyone feels like taking on that challenge…
Written by Kieran Leschinski